insights
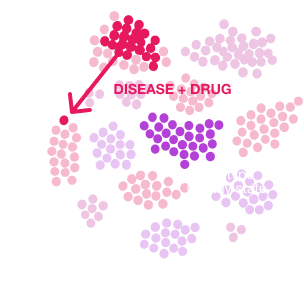
Each dot is a compound. The position of a drug on the plot is called its 'fingerprint'. This fingerprint is the functional identity of the drug, as much as physical measurements (e.g. spectroscopy, NMR, crystallography) uniquely identify a compound.
Compounds that have tightly clustered fingerprints cause neurons to behave indistinguishably. Example cluster: Different Kv7 activator drugs used for epilepsy, pain, ALS.
This 'functional identity' can be used to quickly seek a replacement drug candidate for a chosen compound (screening through optimization).
Pharmas can use this measurement to find a replacement for drugs that are going off patent, find 'work-alikes' or 'electro-mimetics' to competitors' drugs, identify the target of a test compound, or compare the efficacy of their compounds to competitors' drugs.
We have built the world's only perturbation atlas of neuronal behavior to transform the future of neurotherapeutics by guiding medicines to the right targets, validating their efficacy, and ensuring drug safety. Our multimodal dataset is powered by scale: Quiver has now collected a perturbation dataset in human neurons across 18,000 genes and 3,500 compounds.

- 1.5B Action Potentials
- >25M Individual Neurons


- 500+ Features
- Single Cell, Single Action Potential Resolution
- >500k Neuron/Day
- Neuronal Firing and Synaptic Function + Plasticity


- 0.5B Action Potentials
- >6M Individual Neurons

- 1.5B Action Potentials
- >25M Individual Neurons

- 500+ Features
- Single Cell, Single Action Potential Resolution
- >500k Neuron/Day
- Neuronal Firing and Synaptic Function + Plasticity

- 0.5B Action Potentials
- >6M Individual Neurons
Our high-dimensional platform maps how genes, drugs, and diseases perturb neurons at single-cell resolution. Integration of this powerful experimental data with in silico predictions, through our AI agent (QnAi; Quiver's Question and Answer AI), allows us to refine hypotheses, creating a self-improving discovery cycle that accelerates CNS drug development.
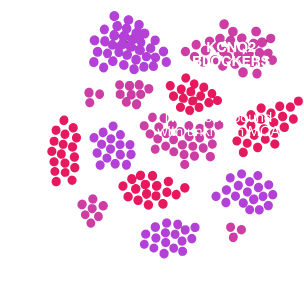
- Target deconvolution: Pinpoint true disease drivers and mechanisms hidden from traditional pipelines.
- Predictive screening & toxicity avoidance: De-risk programs early by predicting efficacy and safety before costly in vivo or clinical studies.
- Therapeutic matching: Align drugs with disease “opposite signatures,” genetic targets, or look-alike compounds to surface new treatments and repurposing opportunities.
- Disease clustering & mechanism discovery: Reveal shared neuronal dysfunction and convergent pathways across disorders.

novel target id
target validation
target deconvolution
Validate Targets


Ranks Lead molecules
phenotypic screens
on-target vector analysis
Phenotype Rescue



in silico prediction
in vivo confirmation
off-vector analysis
Flag Perturbation Risks


novel target id
target validation
target deconvolution
Ranks Lead molecules
phenotypic screens
on-target vector analysis

in silico prediction
in vivo confirmation
off-vector analysis